Report Format – Molecular Gastrointestinal Pathogens (Bacteria)
The RCPAQAP Microbiology has changed the report format issued to participants enrolled in the Molecular Gastrointestinal Pathogens (Bacteria) Program. The new report format follows a standard structure that will be adopted by other programs offered by the RCPAQAP.
The structure of the new report is as follows:
Summary of Performance
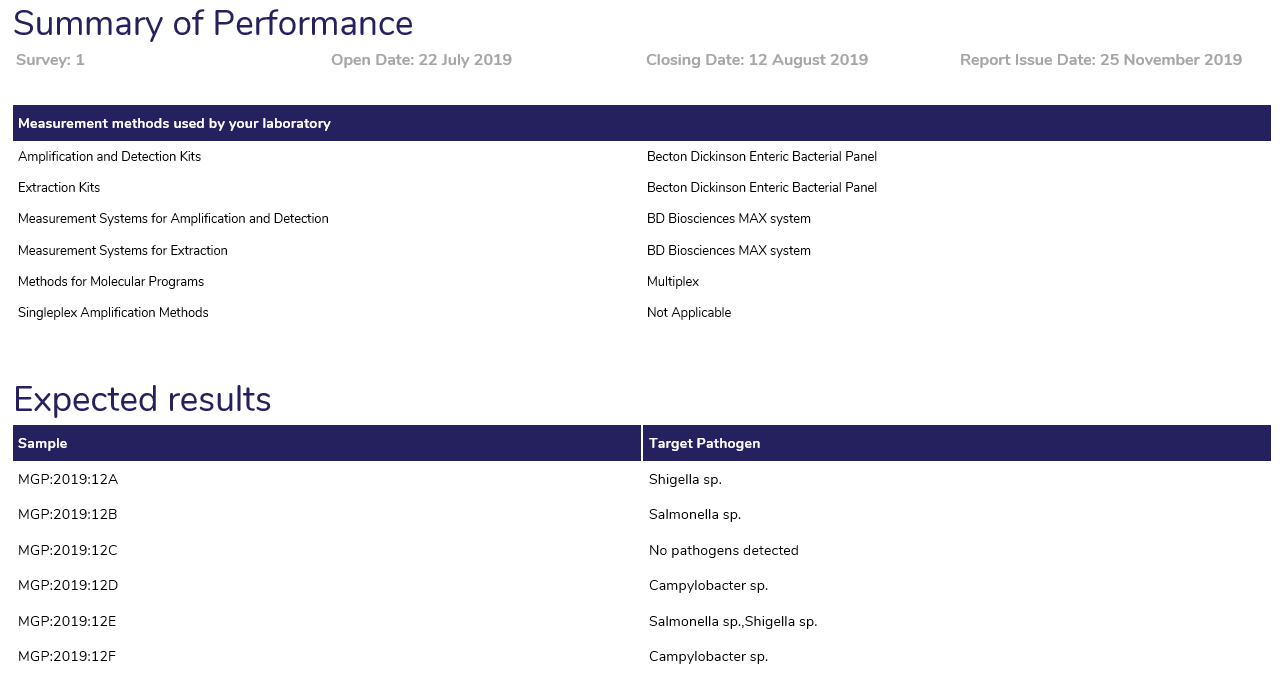
- Method System used by your laboratory: Displays the method supplied by the participant at result entry in the myQAP portal.
- Expected Result: The expected result lists the target pathogens in each sample.
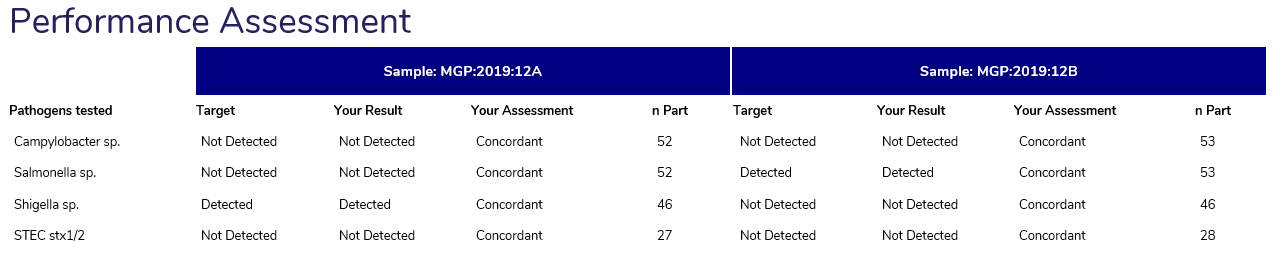
The Performance assessment displays the pathogens tested within the participating laboratory, the target result (if the pathogen is expected to be detected), the participant result, the assessment of the participant’s result (CONCORDANT / DISCORDANT) and the number of participants performing the test.
The Overall Performance provides participants with an indication of the samples that require review if at any time a result does not match the target result.
Result Review
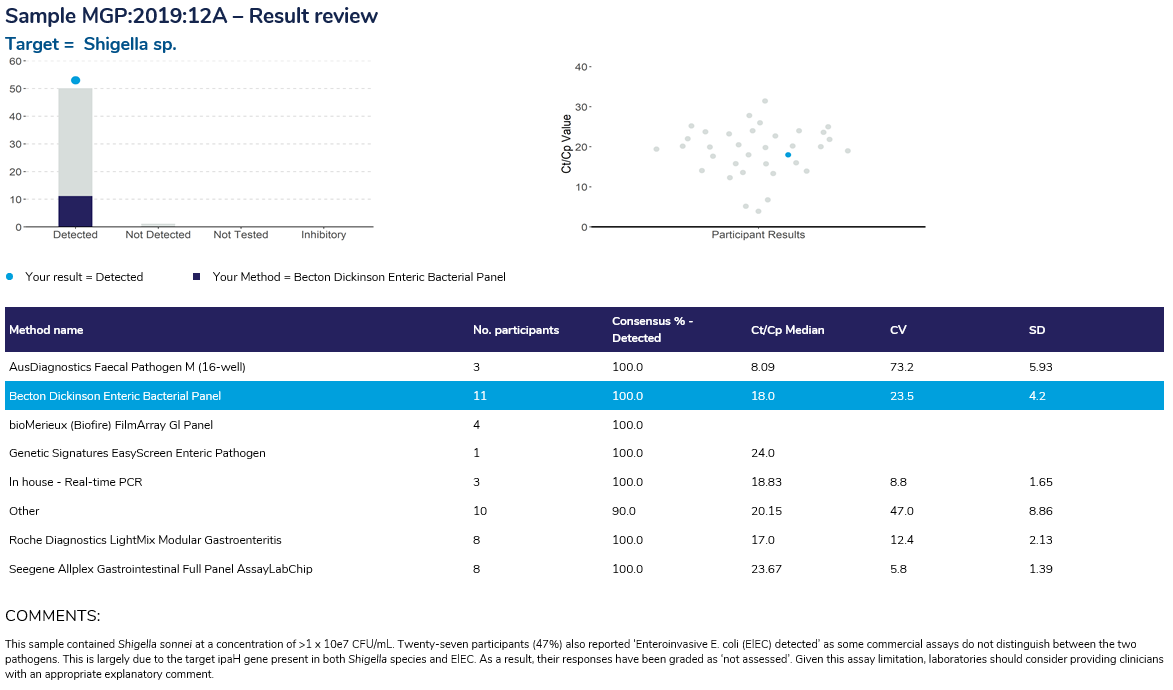
The result review displays the breakdown of the responses received from participants for the target pathogens within each sample. The histogram shows the interpretive comments returned by participants and the scatterplot displays the Ct/Cp value returned, highlighting in blue the result returned by the participant.
The result review page provides a breakdown of the participant results by Amplification / detection kit, displaying the number of participants using the kits, the percentage of participants that returned a result of “Detected” for the pathogen, the median Ct/Cp value, the coefficient of variation (CV) and standard deviation (SD).
Comments are provided for each sample’s result review.
Can't find what you're looking for?

